Services
We offer services in genomics and especially in high-throughput sequencing. We are specialized in functional genomics applications in all eukaryotes, particularly in Transcriptomics: RNA-seq (low input, long-read, single-cell) and Epigenomics/Epitranscriptomics: RNA m6A methylation. Our expertise also lies in long-read sequencing (Nanopore).
We also propose a sequencing service “ready-to-load” for users who make their own libraries.
| Applications | Operating |

Eukaryotic transcriptome (RNA-seq)
We propose a RNA-seq service with short-read Illumina sequencing (NextSeq 2000) and long-read sequencing (Oxford Nanopore Technologies MinION and PromethION).
SHEET (French)
General applications: RNA-seq data can serve to quantify RNA, measure differences in gene expression or isoforms, analyse splicing variants or polymorphisms, assemble de novo transcriptomes (if the genomic sequence is not available or with fragmentary annotation).
Samples: We work with many eukaryotic animal species, whether they are model organisms or not (human, rat, mouse, fish, frog, fly, butterfly, jellyfish, yeast, plants…). Samples are total RNA (we do not perform the step of RNA extraction), which can be obtained from cells (included sorted cells), tissues (included fixed), biopsies…
Proposed services:
- Assistance with experimental design,
- RNA quantification, RNA quality control,
- Library construction (directional and non-directional) for short read Illumina sequencing, coupled with mRNA purification (polyA+) or rRNA depletion (capture of coding and non-coding RNAs) or a protocol for low input mRNA (polyA+),
- Library construction (non-directional) for long read Nanopore sequencing, coupled with mRNA purification (polyA+), included for low input mRNA (polyA+),
- Quality control of libraries, followed by sequencing in single-end or pair-end or long reads (depending on application),
- Included automated bioinformatics analyses (Illumina): demultiplexing, quality control of the reads with report, adapter trimming, read mapping, quantification and differential expression for simple designs (pairwise comparisons),
- Optional bioinformatics analyses (Nanopore): basecalling, demultiplexing, quality control of sequences,
- Deposition of results in public databases (GEO, NCBI) prior to publication,
- Other analyses performed by a bioinformatician: usage of an annotated genome not accessible from the Ensembl database, complex experimental designs, non-standard parameters for the analyses, Nanopore analyses including isoform detection.
Single-cell eukaryotic transcriptome (scRNA-seq)
We propose a scRNA-seq service (10x Genomics) with short read Illumina sequencing (NextSeq 2000) or long read sequencing (Oxford Nanopore Technologies MinION and PromethION) enabling to study isoforms at the single cell level.
SHEET (French)
General applications: scRNA-seq data can serve to explore the heterogeneity of cells within a tissue (including tumors), detect sub-populations or rare cell types.
Samples: We work with many eukaryotic animal species, whether they are model organisms or not (human, rat, mouse, fish, frog, fly, butterfly, jellyfish, yeast, plants…). Samples are either fresh cells dissociated or sorted (only within IBENS), or fixed cells (we are involved in the development of an approach using the ACME protocol).
Proposed services:
- Assistance with experimental design,
- Counseling for sample preparation,
- Users within IBENS : usage of the 10x genomics Chromium by users,
- Users external to IBENS : delivery of fixed samples with ACME protocol,
- Library construction for short read Illumina sequencing, quality control of libraries, followed by pair-end sequencing,
- Library construction for long read Nanopore sequencing,
- Included automated bioinformatics analyses : demultiplexing, quality control of the reads with report,
- Other analyses performed by a bioinformatician: read mapping of public or private genome (possibly including project-specific sequences like fluorescent marker), quantification with annotation, production of the CellRanger report (10x genomics).
Regulome (m6A)
We propose a service to construct the libraries for detection of modified nucleotides on RNA (m6A) with RNA direct sequencing (Nanopore).
General applications m6A: Epitranscriptomics enables to identify and quantify modified nucleotides within RNA. The modified base m6A (N6-methyladenine) is the most common modification in eukaryotic mRNA.
Samples: We work with many eukaryotic animal species, whether they are model organisms or not (human, rat, mouse, fish, frog, fly, butterfly, jellyfish, yeast, plants…). m6A: Samples are total RNA (we do not perform the step of RNA extraction), which can be obtained from cells (included sorted cells), tissues (included fixed), biopsies…
Proposed services:
- Assistance with experimental design,
- Quantification of immunoprecipitated DNA or RNA, quality control of RNA,
- Library construction (non-directional) for long read direct RNA Nanopore sequencing (to detect m6A modifications), coupled with mRNA purification (polyA+), included for low input mRNA (polyA+),
- Quality control of the libraries,
- Deposition of results in public databases (GEO, NCBI) prior to publication,
- Other analyses performed by a bioinformatician: removal of low-quality reads, detection of m6A modifications.
Service overview
I) Library preparation
Our genomcis core facility offers the following library preparation protocols:
| Short read library protocols (Illumina) |
Long read library protocols (Oxford Nanopore Technologies) |
|
|---|---|---|
| RNA-seq | PolyA mRNA purification rRNA depletion (long non-coding RNAs) |
PolyA mRNA purification |
| RNA-seq low input | Low input amplification | Low input amplification |
| 10x Genomics 3’ Gene Expression | 10x Genomics 3’ Gene Expression (scNauMi-seq protocol) |
|
| Regulome |
N/A |
Direct RNA (m6A modification detection) |
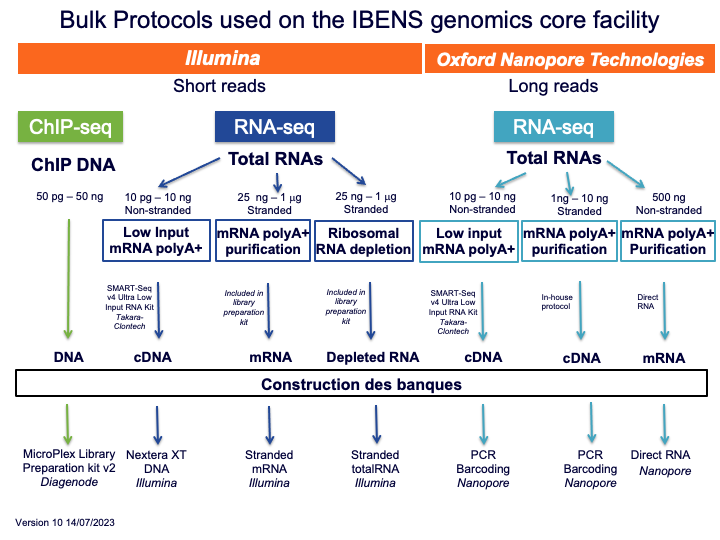
The following schema allow to choose the protocol against the biological question:
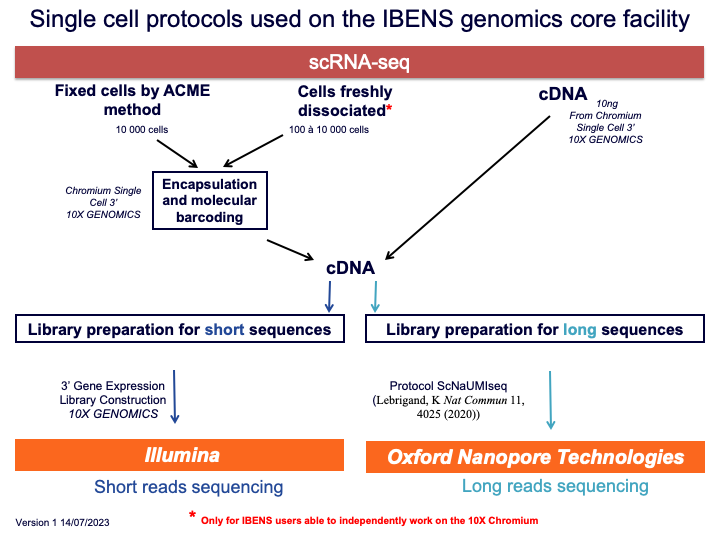
Here all the single-cell protocols available on our genomics core facility:
II) Sequencing
Our genomics core facility have sequencers from two supplier: Illumina for short reads sequencing and Oxford Nanopore Technologies for long reads sequencing.
We also propose a sequencing service “ready-to-load” for users who make their own libraries.
Short reads sequencing
Illumina sequencers allow to perform single and or paired-end sequencing. With paired-end sequencing, the two ends of the inserts between adapters are sequenced instead of just one in single end mode. In most the the cases, the length of the two ends are the sames (they are commonly noted like 2 × 100).
The GenomiqueENS core facility currenly use an Illumina NextSeq 2000 sequencer for short reads sequencing.
| Illumina NextSeq 2000 | |||||||
|---|---|---|---|---|---|---|---|
| Flowcell P1 (~100 million reads per run) |
Flowcell P2 (~400 million reads per run) |
Flowcell P3 (~1.2 billion reads per run) |
Flowcell P4 (~1.8 billion reads per run) |
||||
| Kit | Size (bp) | Kit | Size (bp) | Kit | Size (bp) | Kit | Size (bp) |
| N/A | N/A | N/A | 50 cycles | 1 × 50 2 × 25 |
|||
| 100 cycles | 1 × 100 | 100 cycles | 1 × 100 2 × 50 |
100 cycles | 1 × 100 2 × 50 |
100 cycles | 1 × 100 2 × 50 |
| N/A | 200 cycles | 2 × 100 | 200 cycles | 2 × 100 | 200 cycles | 2 × 100 | |
| 300 cycles | 2 × 150 | 300 cycles | 2 × 150 | 300 cycles | 2 × 150 | 300 cycles | 2 × 150 |
| 600 cycles | 2 × 300 (*) | 600 cycles (*) | 2 × 300 | N/A | N/A | ||
Long reads seqencing
Oxford Nanopore Technologies sequencers allow to sequence DNA or RNA. The read lengths and the throughput vary against the sequencing mode:
| Oxford Nanopore Technologies MinION and PromethION P2 | |||
|---|---|---|---|
| Kit | Size (bp) | MinION read count |
PromethION read count |
| DNA (cDNA) |
Until several thousands kb (full length transcripts in RNA-seq) |
6–8 million reads | 80–120 million reads |
| Direct RNA | ~1,000 bp mean size | ~1 million reads | Under testing |
III) Bioinformatics analyses
Our genomics core facility offers two service levels for bioinformatics analysis (the descriptions of the bioinformatics services are available in the application sections of this page):
- Automated analysis included in the sequencing price, without the help of a bioinformatican: basecalling, demultiplexing, quality control of the reads, and more for some application type
- An optional accompanied analysis, with the help of a bioinformatician
Bioinformatics analyses are only performed on data sequenced by our genomics core facility. To analyse your data, you can contact a bioinformatics core facility.
Proceedings of a collabirating project with our genomics core facility
Since 2006, we have created an “accompanied project” service as a scientific collaboration that handle all the experimental process and provide “read-to-publish results”. You benefit from our experience and our know-how from experimental design to the data analysis and an help to publication writing.
We are open to academic laboratories and companies. Most of our project are performed with teams outside the IBENS.
SERVICE SHEET
Publications: affiliations and acknowledgments to mention
Research collaboration implies that the two persons working on experiments and bioinformatics associated to the project are listed as co-authors the first time the results are published. This is only the case if the core facility has been involved in library preparation and/or bioinformatics analysis (except automated analyses), not in the case of “ready-to-load” projects. The affiliation to use is the following:
GenomiqueENS, Institut de Biologie de l'ENS (IBENS), Département de biologie, École normale supérieure, CNRS, INSERM, Université PSL, 75005 Paris, France
For all projects (including “ready-to-load” projects), reference to the "France Genomique” infrastructure must appear in the acknowledgements section of the article, with the following sentence:
The GenomiqueENS core facility was supported by the France Génomique national infrastructure, funded as part of the "Investissements d'Avenir" program managed by the Agence Nationale de la Recherche (contract ANR-10-INBS-09)
